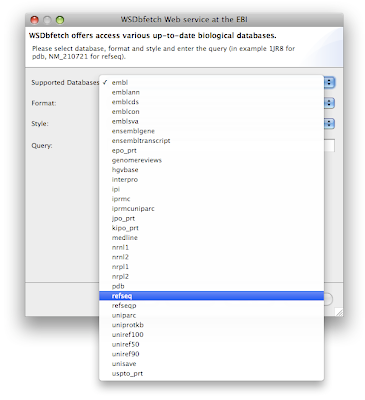
On behalf of all
Bioclipse developers I am happy to announce the release of Bioclipse 2.0. Bioclipse is a free, open source workbench for the life sciences that provides advanced functionality mainly in cheminformatics (bioinformatics is planned for version 2.1 later this summer). Some major components include a brand new chemical editor for SWT (JChemPaint), interactive 3D visualization of molecules (
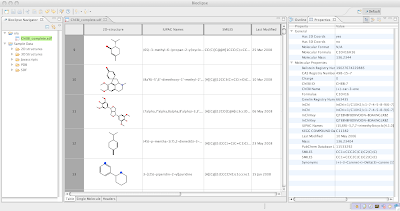
Jmol), a Molecules Table
capable of reading large files, and a powerful backbone in cheminformatics provided by the
Chemistry Development Kit (CDK) library.
 Figure 1: Screenshot of Bioclipse showing editing of a chemical structure using the new JChemPaint editor.
Figure 1: Screenshot of Bioclipse showing editing of a chemical structure using the new JChemPaint editor. Bioclipse is a
Rich Client for the life sciences that provides the means to run and integrate algorithms and tools in disconnected state, while still taking advantage of remote services if a network connection is available. Built on the famous
Eclipse framework, Bioclipse delivers a state-of-the-art plugin architecture which makes it possible to extend it in any direction.
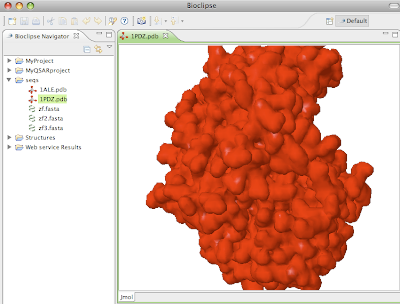
 Figure 2: Screenshot of the interactive 3D visualization of a protein using the integrated component Jmol.
Figure 2: Screenshot of the interactive 3D visualization of a protein using the integrated component Jmol. All functionality in Bioclipse 2 is available from the GUI as well as a new scripting language based on Javascript. This allows for complete control of the workbench and functionality from scripts, which can be used to automate tasks or reproduce and validate scientific analyses.
 Figure 3: All functionality in Bioclipse is available from an integrated scripting language based on Javascript.
Figure 3: All functionality in Bioclipse is available from an integrated scripting language based on Javascript. Bioclipse 2 can be downloaded from
Sourceforge, releases are available for all major platforms. There is an update site where users can install additional functionality (such as
Speclipse) and data collections; this is available from the Bioclipse workbench under menu
Help > Software updates.
A small
installation guide is also provided, but the main documentation for Bioclipse is available from
help.bioclipse.net; the same information is also available from within Bioclipse from the menu
Help > Help Contents. For general questions there is the
bioclipse-users mailing list.
All software contains bugs, and Bioclipse is no exception. However, in contrast to many commercial and closed source initiatives, open source projects generally have a faster bug fixing rate as well as more frequent releases. If you find bugs in Bioclipse, please report them on
bugs.bioclipse.net. There is a list of
intractable bugs on the
Bioclipse development wiki, and also a convenience list for tracking
known major bugs.
Bioclipse is an open development that welcome new developers with varying backgrounds. Developers hang out on daily basis on IRC (irc.freenode.net, channel
#bioclipse), and can also be reached via the mailing list
bioclipse-devel.
Thanks to all contributors who made this release possible!