ws.downloadDbEntry(String db, String query, String format)
From the GUI you can use menu: File > New... and select the Wizard Download > Query WSDbfetch at EBI. Click next and see the many available databases in the dropdown list Supported databases:
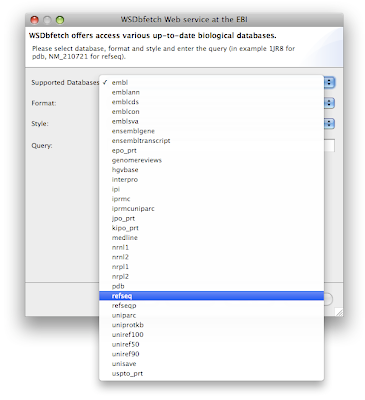 Try for example to download the sequence "NM_210721" from the "refseq" database.
Try for example to download the sequence "NM_210721" from the "refseq" database.If you'd like to download proteins in PDB format, there is a convenience wizard available in the New Wizards dialog available from the menu File > New...
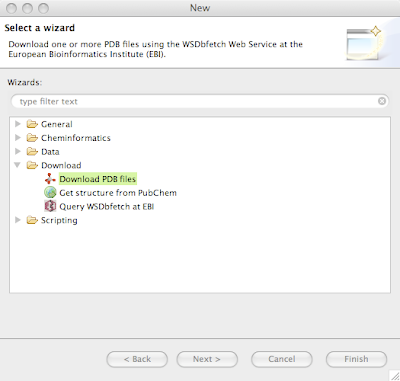
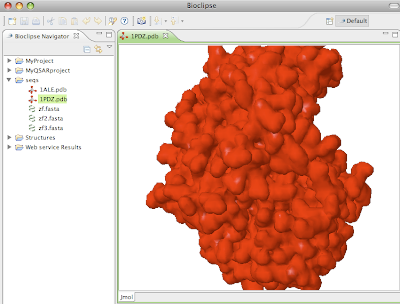
Clicking Next allows for inputting a comma-separated list of PDB IDs, try for example "1ale,2pdz". Clicking next downloads the file to the selected folder in the Navigator, or to the Virtual project if nothing is selected. Below is visualized 2PDZ in the Jmol Editor.



No comments:
Post a Comment