The Bioclipse team is proud to announce the release
of Bioclipse 1.0.
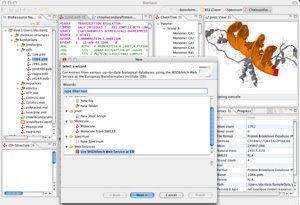
Bioclipse is a free, open source, workbench for chemo- and bioinformatics with powerful editing and visualization capabilities for molecules, sequences, proteins, spectra etc. The major features of version 1.0 are:
* Import and export in various file formats
* Visual editing of molecular 2D-structures
* 3D-visualization of molecules and proteins
* Editing and visualization of sequences and features (DNA, RNA, proteins etc)
* Graphing and editing of various types of spectra, e. g. NMR, MS
* Retrieval of resources (sequences, proteins, etc) from public data repositories
* Scripting of 3D-visualizations with syntax highlighting and content assistance
* PDB-editor with syntax highlighting for working with PDB files
* CMLRSS-viewer for downloading chemical content published on the web using RSS-feeds
* Integrated, searchable help-system
* Hierarchical view of molecular and macromolecular substructures and calculation of chemical properties* Connection with external programs, e. g. PyMol
Bioclipse is a rich client, which means it is run on your local computer but also gives the possibility to communicate with servers for data retrieval and computational services. The powerful plugin architecture is based on Eclipse, and results in a responsive, integrated user interface designed for simple and intuitive operations that at the same time is easy to extend with custom functionality.
There is much ongoing work with Bioclipse and new features are constantly added. Please visit the Bioclipse Wiki in order to get the latest information regarding the development.
Bioclipse is available for download from Sourceforge.
Thanks to all developers who have made this release possible!


